Papers: Independent lab
2023:
Armendariz DA*, Goetsch SC*, Sundarrajan A, Sivakumar S, Wang Y, Xie S, Munshi NV#, Hon GC#. CHD-associated enhancers shape human cardiomyocyte lineage commitment. Elife. 2023.
Cooley A*, Madhukaran S*, Stroebele E, Colon Caraballo M, Wang L, Akgul Y, Hon GC#, Mahendroo M#. Dynamic states of cervical epithelia during pregnancy and epithelial barrier disruption. iScience. 2023.
Bhattacharyya S, Kollipara RK, Orquera-Tornakian G, Goetsch S, Zhang M, Perry C, Li B, Shelton JM, Bhakta M, Duan J, Xie Y, Xiao G, Evers BM, Hon GC, Kittler R, Munshi NV. Global chromatin landscapes identify candidate noncoding modifiers of cardiac rhythm. J Clin Invest. 2023.
2022:
Bhattacharyya S*, Duan J*, Vela RJ, Bhakta M, Bajona P, Mammen PPA, Hon GC#, Munshi NV#. Accurate Classification of Cardiomyopathy Diagnosis by Chromatin Accessibility. Circulation. 2022.
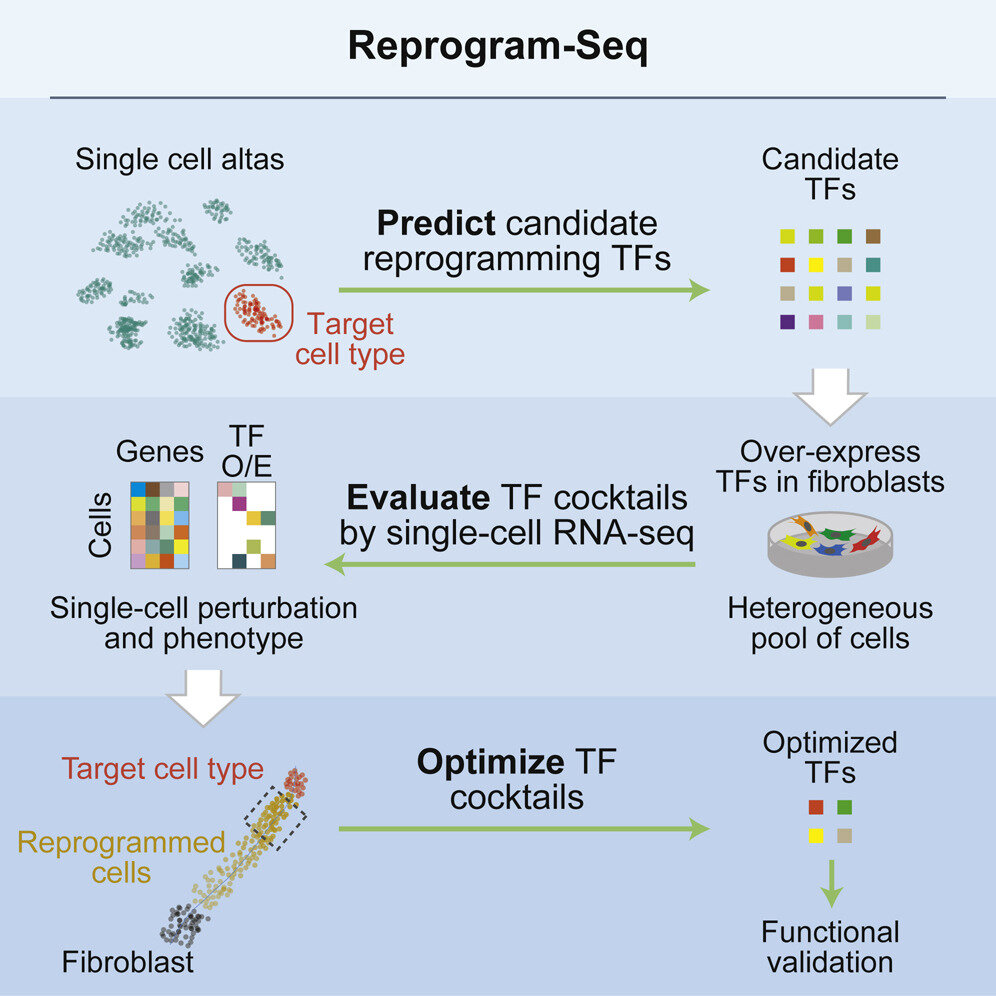
Zhang Y*, Li B*, Cananzi S, Han C, Wang L, Zou Y, Fu Y, Hon GC#, Zhang C#. A single factor elicits multilineage reprogramming of astrocytes in the adult mouse striatum. PNAS. 2022.
Wang Y, Xie S, Armendariz D, Hon GC. Computational identification of clonal cells in single-cell CRISPR screens. BMC Genomics. 2022.
2021:
Yu L*, Wei Y*, Duan J*, Schmitz DA, Sakurai M, Wang L, Wang K, Zhao S, Hon GC#, Wu J#. Blastocyst-like structures generated from human pluripotent stem cells. Nature. 2021.
Duan J, Hon GC. FBA: feature barcoding analysis for single cell RNA-Seq. Bioinformatics. 2021.
Li B, Hon GC. Single-Cell Genomics: Catalyst for Cell Fate Engineering. Front Bioeng Biotechnol. 2021.
2020:
England AR, Chaney CP, Das A, Patel M, Malewska A, Armendariz D, Hon GC, Strand DW, Drake KA, Carroll TJ. Identification and characterization of cellular heterogeneity within the developing renal interstitium. Development. 2020.
2019:
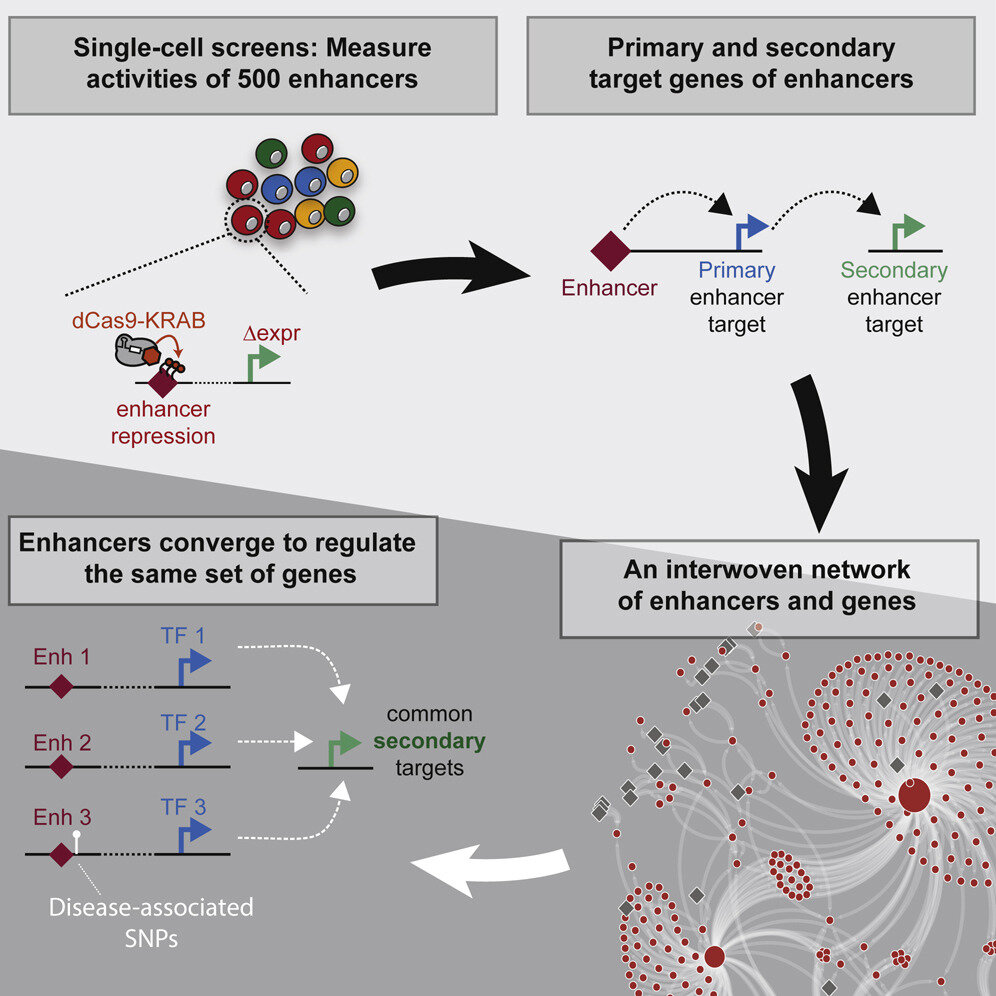
Xie S, Armendariz D, Zhou P, Duan J, Hon GC. Global analysis of enhancer targets reveals convergent enhancer-driven regulatory modules. Cell Reports. 2019.
Zhang Z, Luo D, Zhong X, Choi JH, Ma Y, Wang S, Mahrt E, Guo W, Stawiski EW, Modrusan Z, Seshagiri S, Kapur P, Hon GC, Brugarolas J, and Wang T. SCINA: A Semi-Supervised Subtyping Algorithm of Single Cells and Bulk Samples. Genes. 2019.
Duan J*, Li B*, Bhakta M*, Xie S, Zhou P, Munshi NV‡, Hon GC‡. Rational reprogramming of cellular states by combinatorial perturbation. Cell Reports 2019.
Bhattacharyya S, Duan J, Wang L, Li B, Bhakta M, Fernandez-Perez A, Hon GC, Munshi NV. Using Gjd3-CreEGFP mice to examine atrioventricular node morphology and composition. Sci Rep. 2019.
Xie S, Hon GC. Experimental and Computational Approaches for Single-Cell Enhancer Perturbation Assays. Computational Methods for Single-Cell Data Analysis 2019.
2018
Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell Rep. 2018.
Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, Gupta RK. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018.
Xie S*, Cooley A*, Armendariz D, Zhou P, Hon GC. Frequent sgRNA-barcode recombination in single-cell perturbation assays. PLoS ONE 2018.
2017
Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, Dickerson KE, Xie S, Hon GC, Xuan Z, Zhang MQ, Shao Z, Xu J. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell. 2017.
Partin AC, Ngo TD, Herrell E, Jeong BC, Hon G, Nam Y. Heme enables proper positioning of Drosha and DGCR8 on primary microRNAs. Nat Commun 2017.
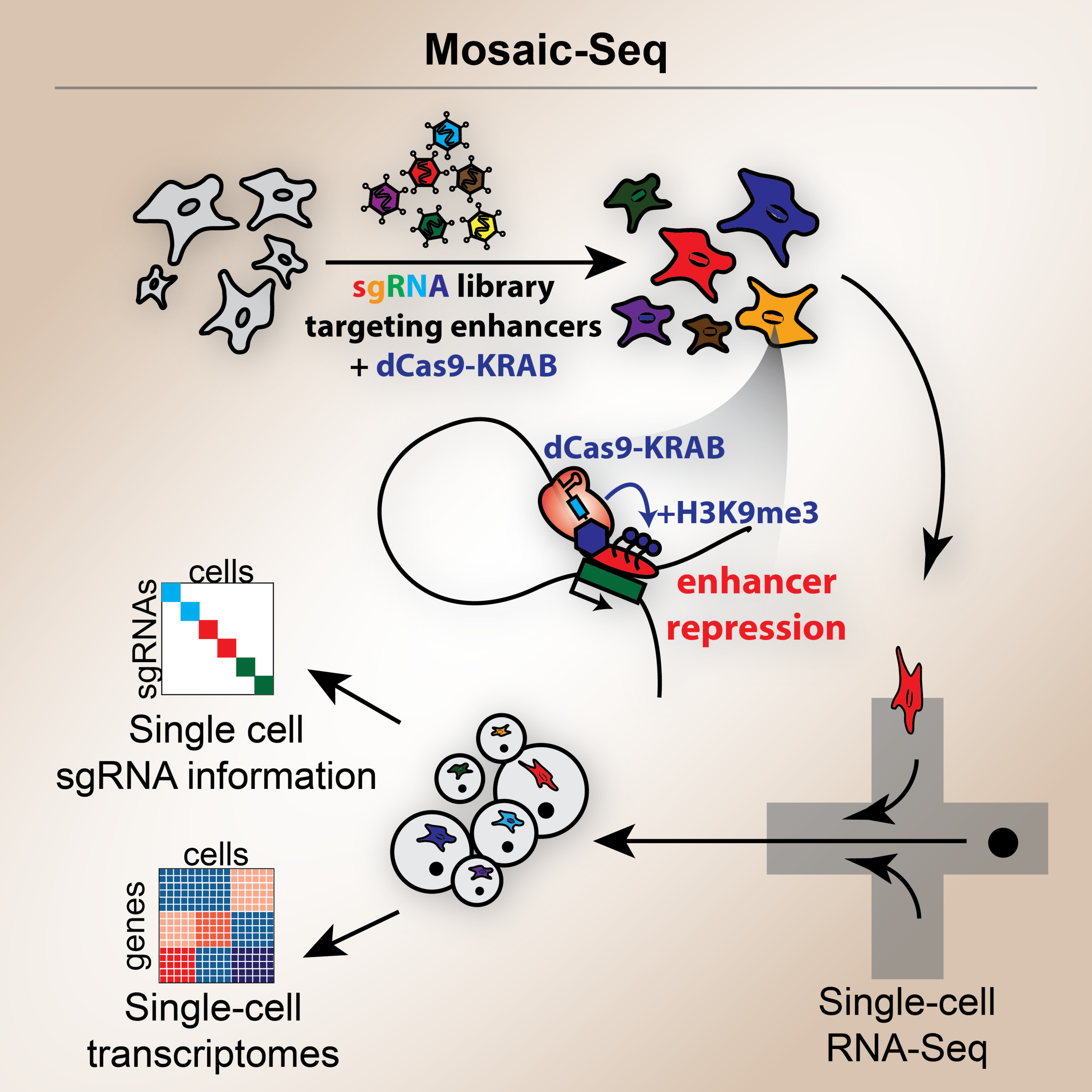
Xie S, Duan J, Li B, Zhou P, Hon GC. Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol Cell 2017.
Papers: Before independent lab
Yu M, Han D, Hon GC, He C. Tet-Assisted Bisulfite Sequencing (TAB-seq). Methods Mol Biol. 2018.
Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, Gu Y, Lum KM, Masui K, Yang H, Rong X, Hong C, Turner KM, Liu F, Hon GC, Jenkins D, Martini M, Armando AM, Quehenberger O, Cloughesy TF, Furnari FB, Cavenee WK, Tontonoz P, Gahman TC, Shiau AK, Cravatt BF, Mischel PS. An LXR-Cholesterol Axis Creates a Metabolic Co-Dependency for Brain Cancers. Cancer Cell. 2016.
Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, Ye Z, Li B, Kuan S, Lee AY, Zanca C, Wei B, Lucey G, Jenkins D, Zhang W, Barr CL, Furnari FB, Cloughesy TF, Yong WH, Gahman TC, Shiau AK, Cavenee WK, Ren B, Mischel PS. EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell 2015
Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, Kuan S, Edsall LE, Zhao BS, Xu GL, He C, Ren B. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014.
Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013.
Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, Nery JR, Qiao H, Yang A, Jamali A, Chen H, Ideker T, Ren B, Bar-Joseph Z, Hughes TR, Ecker JR. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife. 2013.
Zhang L, Szulwach KE, Hon GC, Song CX, Park B, Yu M, Lu X, Dai Q, Wang X, Street CR, Tan H, Min JH, Ren B, Jin P, He C. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Commun. 2013.
Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012.
Smallwood A, Hon GC, Jin F, Henry RE, Espinosa JM, Ren B. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012.
Yu M*, Hon GC*, Szulwach KE*, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012.
Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, Camargo AA, Stevenson BJ, Ecker JR, Bafna V, Strausberg RL, Simpson AJ, Ren B. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012.
Hawkins RD*, Hon GC*, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, Kuan S, Yu P, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Ren B. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011.
Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011.
Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010.
Hawkins RD*, Hon GC*, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010.
Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009.
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009.
Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009.
Heintzman ND*, Hon GC*, Hawkins RD*, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009.
Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput Biol. 2008.
ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007.
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007.
Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006.
Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004.
Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004.